Page under construction
Estimating cardiomyofiber strain in vivo by solving a computational model
L.E. Perotti*, I.A. Verzhbinsky*, K. Moulin, T.E. Cork, M. Loecher, D. Balzani, and D.B. Ennis (2020)
Since heart contraction results from the electrically activated contraction of millions of cardiomyocytes, a measure of cardiomyocyte shortening mechanistically underlies cardiac contraction. In this work we aim to measure preferential aggregate cardiomyocyte (“myofiber”) strains based on Magnetic Resonance Imaging (MRI) data acquired to measure both voxel-wise displacements through systole and myofiber orientation. In order to reduce the effect of experimental noise on the computed myofiber strains, we recast the strains calculation as the solution of a boundary value problem (BVP). This approach does not require a calibrated material model, and consequently is independent of specific myocardial material properties. The solution to this auxiliary BVP is the displacement field corresponding to assigned values of myofiber strains. The actual myofiber strains are then determined by minimizing the difference between computed and measured displacements. The approach is validated using an analytical phantom, for which the ground-truth solution is known. The method is applied to compute myofiber strains using in vivo displacement and myofiber MRI data acquired in a mid-ventricular left ventricle section in N=8 swine subjects. The proposed method shows a more physiological distribution of myofiber strains compared to standard approaches that directly differentiate the displacement field.
More information about this project can be found here.
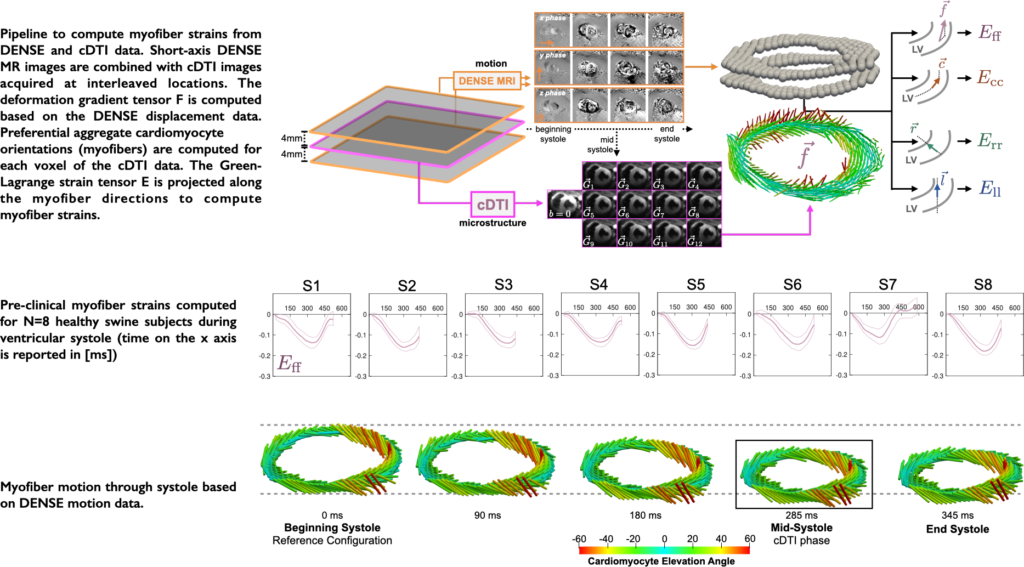
Evaluating myofiber strains using in vivo diffusion and displacement encoded MRI
I.A. Verzhbinsky*, L.E. Perotti*, K. Moulin, T.E. Cork, M. Loecher, and D.B. Ennis (2019)
Aggregate cardiomyocyte (myofiber) shortening and relaxation drive global left ventricular contraction and filling. As such, in vivo myofiber strain (Eff) has mechanistic significance, although currently there is no established method to measure Eff.
The objective of this work is to describe and validate a pipeline to compute Eff based on in vivo MRI data. Our pipeline integrates LV motion from multi-slice Displacement Encoded with Stimulated Echoes (DENSE) MRI with in vivo LV microstructure from cardiac Diffusion Tensor Imaging (cDTI) data. The proposed pipeline is validated using an analytical deforming heart-like phantom. The phantom is used to evaluate the computed 3D cardiac strains.
The validated framework is applied to experimental DENSE MRI and cDTI data acquired in eight (N = 8) healthy swine. The experimental study demonstrated that Eff has decreased transmural variability compared to radial and circumferential strains. The spatial uniformity and mechanistic significance of in vivo Eff make it a compelling candidate for characterization and early detection of cardiac dysfunction.
More information about this project can be found here.